Code
library(data.table)
library(ggplot2)
library(stringr)
library(ggpubr)
library(cowplot)
library(dplyr)HCPcov = fread(paste0(HCPdir,list.files(path = HCPdir, pattern = "unrestricted_annafurtjes")))
HCPcov = HCPcov[,c("Subject", "Release", "Acquisition","fMRI_3T_ReconVrs")]
HCP = fread(paste0(HCPdir,"unrestricted_hcp_freesurfer.csv"))
HCP = HCP[,c("Subject", "Gender", "FS_InterCranial_Vol", "FS_BrainSeg_Vol_No_Vent")]
HCP = merge(HCP, HCPcov, by = "Subject")
HCPage = fread(paste0(HCPdir, "RESTRICTED_annafurtjes_12_14_2023_4_18_2.csv"))
HCPage = HCPage[,c("Subject","Age_in_Yrs", "Family_ID")]
HCP = merge(HCP, HCPage, by ="Subject")
names(HCP) = c("ID", "Sex", "ICV", "TBV", "Release", "Acquisition","fMRI_3T_ReconVrs", "Age", "Family_ID")
# convert mm3 estimates to more intuitive cm3 estimates
HCP$ICV = HCP$ICV/1000
HCP$TBV = HCP$TBV/1000
# calculate atrophy scores
HCP$diff = HCP$ICV - HCP$TBV
HCP$ratio = HCP$TBV / HCP$ICV
# delete those from data
HCP=HCP[-which(HCP$diff < 0),]
# estimate residual model
model <- lm(TBV ~ ICV, data = HCP)
HCP$resid = as.vector(resid(model, na.rm=T))
# standardise variables
HCP$diff_stand = as.vector(scale(HCP$diff))
HCP$ratio_stand = as.vector(scale(HCP$ratio))
HCP$resid_stand = as.vector(scale(HCP$resid))It is contrary to our expectation that ICV gets so much larger with younger age - cohort effect?
Much stronger correlation between younger age and ICV than there should be (N = 1103)
This explains why the atrophy scores with dependence of baseline levels are also strongly affected by age
The only score that is independent of baseline levels - i.e., residual score - has no association with age
corICV = ggplot(data = HCP, aes(x = ICV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
corTBV = ggplot(data = HCP, aes(x = TBV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
cordiff = ggplot(data = HCP, aes(x = diff, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Difference score")+
theme(panel.border = element_blank())
corratio = ggplot(data = HCP, aes(x = ratio, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Ratio score")+
theme(panel.border = element_blank())
corresid = ggplot(data = HCP, aes(x = resid, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Residual score")+
theme(panel.border = element_blank())
ggarrange(corICV, corTBV, cordiff, corratio, corresid, common.legend=T, legend = "bottom")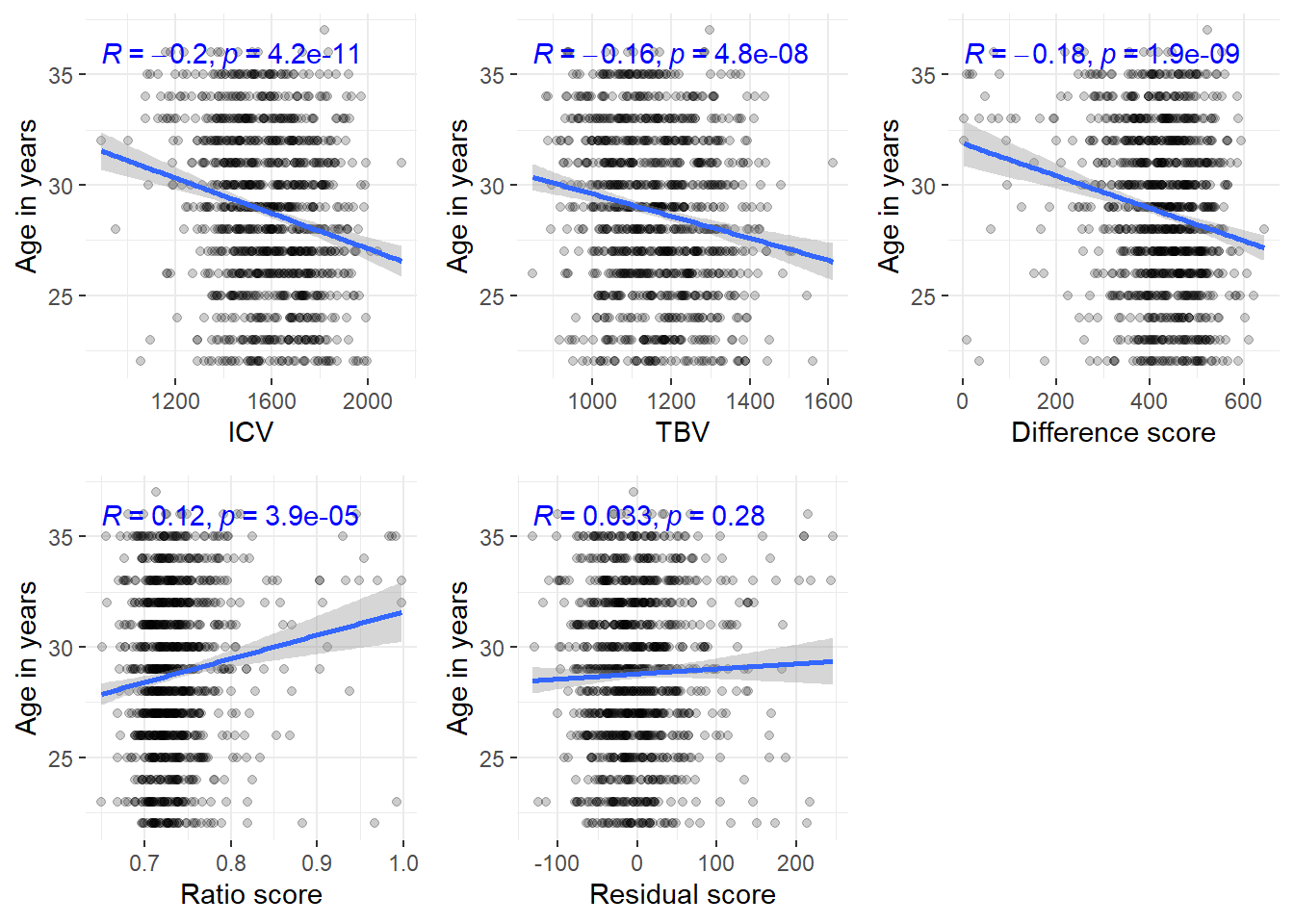
Relatedness is determined based on Family_ID variable
The correlations stay about the same even when excluding all related individuals (N = 444)
##### Family_ID
## Randomly select one participant from each family and keep only one
HCPunrel = HCP[, .SD[sample(x = .N, size = 1)], by = Family_ID]
# calculate atrophy scores
HCPunrel$diff = HCPunrel$ICV - HCPunrel$TBV
HCPunrel$ratio = HCPunrel$TBV / HCPunrel$ICV
# delete those from data (none in this unrelated data)
if(sum(HCPunrel$diff < 0) != 0){HCPunrel=HCPunrel[-which(HCPunrel$diff < 0),]}
# estimate residual model
model <- lm(TBV ~ ICV, data = HCPunrel)
HCPunrel$resid = as.vector(resid(model, na.rm=T))
# standardise variables
HCPunrel$diff_stand = as.vector(scale(HCPunrel$diff))
HCPunrel$ratio_stand = as.vector(scale(HCPunrel$ratio))
HCPunrel$resid_stand = as.vector(scale(HCPunrel$resid))
########## plot age correlations (copied from Aim 1)
corICVunrel = ggplot(data = HCPunrel, aes(x = ICV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
corTBVunrel = ggplot(data = HCPunrel, aes(x = TBV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
cordiffunrel = ggplot(data = HCPunrel, aes(x = diff, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Difference score")+
theme(panel.border = element_blank())
corratiounrel = ggplot(data = HCPunrel, aes(x = ratio, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Ratio score")+
theme(panel.border = element_blank())
corresidunrel = ggplot(data = HCPunrel, aes(x = resid, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Residual score")+
theme(panel.border = element_blank())
ggarrange(corICVunrel, corTBVunrel, cordiffunrel, corratiounrel, corresidunrel, common.legend=T, legend = "bottom")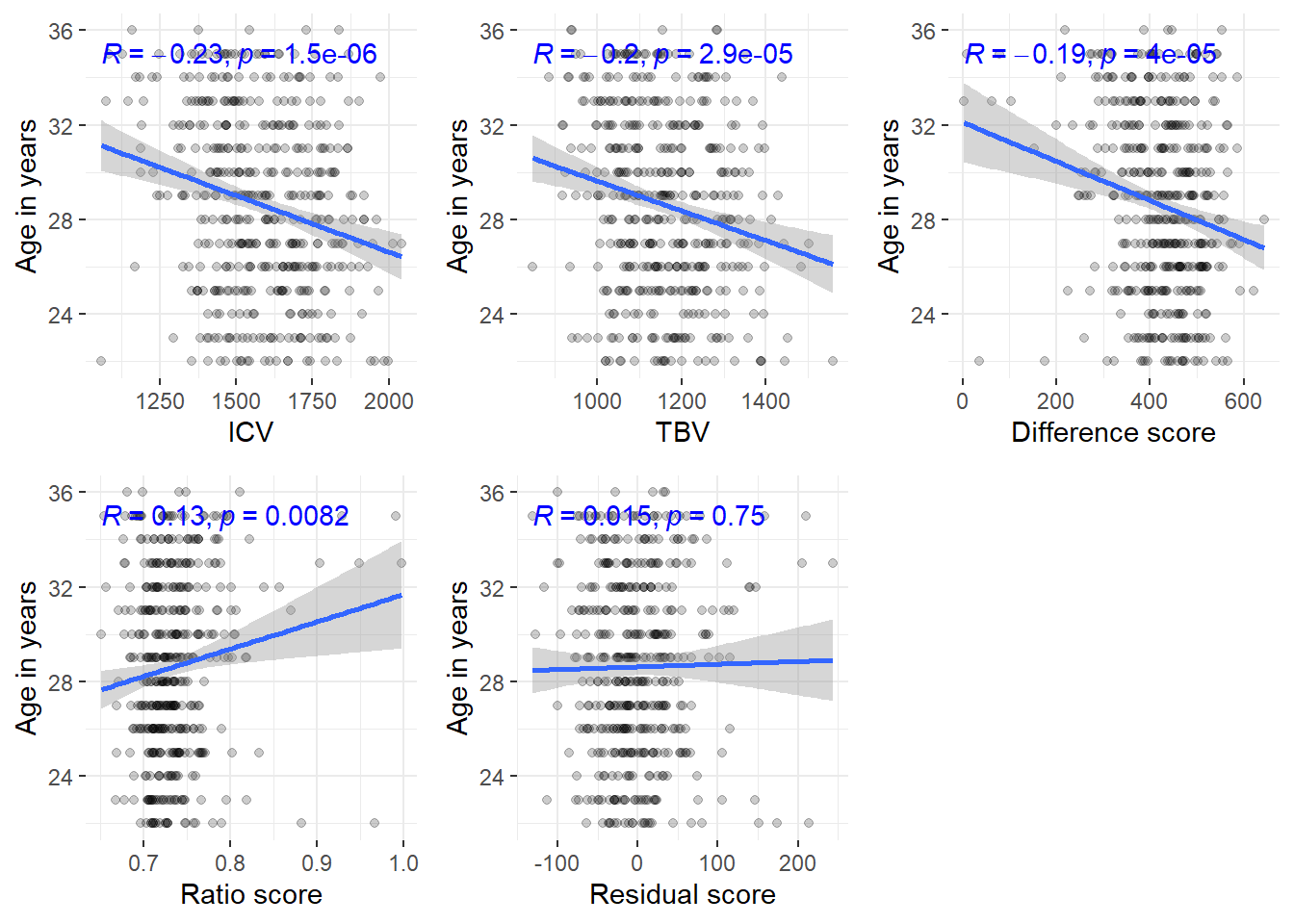
Adjusting for batch effects does not change the strong age correlation we see in this young sample either.
Call:
lm(formula = "ICV ~ Release + Acquisition + fMRI_3T_ReconVrs",
data = HCP)
Residuals:
Min 1Q Median 3Q Max
-658.59 -118.49 0.97 129.22 550.84
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1519.335 93.893 16.182 <2e-16 ***
ReleaseQ1 -40.029 75.514 -0.530 0.596
ReleaseQ2 -47.039 66.367 -0.709 0.479
ReleaseQ3 -47.600 59.833 -0.796 0.426
ReleaseS1200 32.588 54.523 0.598 0.550
ReleaseS500 13.447 55.737 0.241 0.809
ReleaseS900 -18.843 51.431 -0.366 0.714
AcquisitionQ02 57.604 44.320 1.300 0.194
AcquisitionQ03 45.814 59.384 0.771 0.441
AcquisitionQ04 17.944 81.444 0.220 0.826
AcquisitionQ05 5.692 83.162 0.068 0.945
AcquisitionQ06 9.911 82.416 0.120 0.904
AcquisitionQ07 43.858 80.984 0.542 0.588
AcquisitionQ08 49.088 80.722 0.608 0.543
AcquisitionQ09 57.448 81.066 0.709 0.479
AcquisitionQ10 69.126 81.264 0.851 0.395
AcquisitionQ11 102.821 81.136 1.267 0.205
AcquisitionQ12 25.045 82.963 0.302 0.763
AcquisitionQ13 -29.149 83.926 -0.347 0.728
fMRI_3T_ReconVrsr177 37.644 61.707 0.610 0.542
fMRI_3T_ReconVrsr177 r227 30.217 91.884 0.329 0.742
fMRI_3T_ReconVrsr227 34.411 52.511 0.655 0.512
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 180.9 on 1081 degrees of freedom
Multiple R-squared: 0.03469, Adjusted R-squared: 0.01594
F-statistic: 1.85 on 21 and 1081 DF, p-value: 0.01124HCP$ICVadj = resid(model)
corICV_adj = ggplot(data = HCP, aes(x = ICVadj, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("ICV adjusted for Release and Acquisition")+
theme(panel.border = element_blank())
# adjust TBV
model = lm('TBV ~ Release + Acquisition', data =HCP)
HCP$TBVadj = resid(model)
corTBV_adj = ggplot(data = HCP, aes(x = TBVadj, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("TBV adjusted for Release and Acquisition")+
theme(panel.border = element_blank())
# re-create diff score with adjusted variables
HCP$diff_adj = HCP$ICVadj - HCP$TBVadj
cordiff_adj = ggplot(data = HCP, aes(x = diff_adj, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Difference score")+
theme(panel.border = element_blank())
# re-create ratio score with adjusted variables
HCP$ratio_adj = HCP$TBVadj / HCP$ICVadj
corratio_adj = ggplot(data = HCP, aes(x = ratio_adj, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Ratio score")+
theme(panel.border = element_blank())
ggarrange(corICV_adj, corTBV_adj, common.legend = T, legend = "bottom")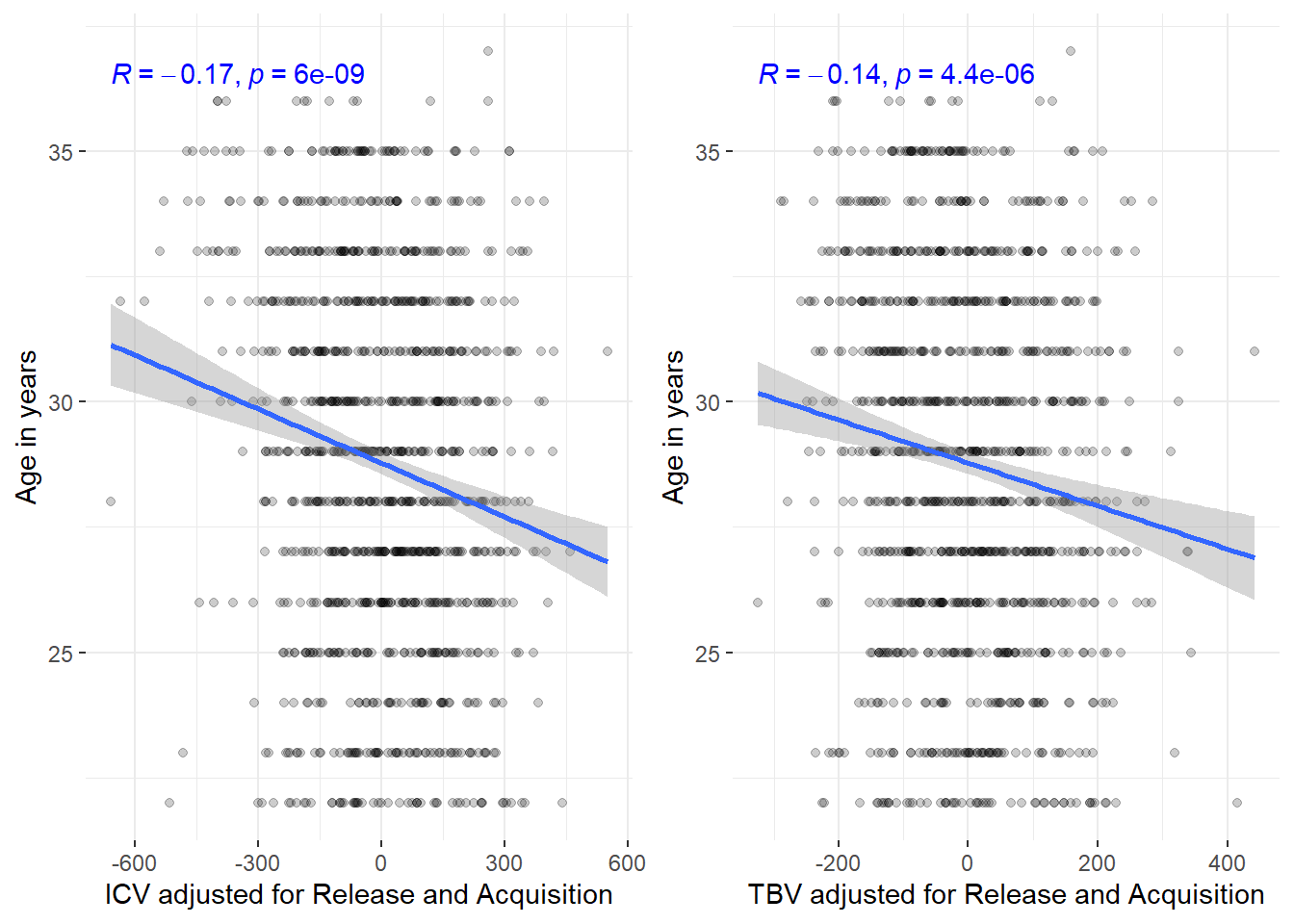
If we consider participants of and below 29 years only, the ICV age correlation disappears
This threshold was empirically determined and then applied in the plot below
In this younger sample, we see no age correlations whatsoever anymore
the plot shows that the ICV-age correlation disappears at age 29 and below
also this sample includes only 3 out of the 10 participants that had to get excluded due to ICV < TBV
### determne which age is a good one to use for cut-off
# determine age cut-offs to iterate through
ageCut = seq(from = min(HCP$Age)+1, to = max(HCP$Age), by = 1)
# object to store results
storeNames = c("Age cut-off value", "Cor", "p", "ci_l", "ci_u", "Measure")
store = as.data.frame(matrix(nrow = length(ageCut), ncol = length(storeNames)))
names(store) = storeNames
# iterate over each age cut-off and calculate scores
for(i in ageCut){
# store which age cut off iteration this is
loc = which(is.na(store$`Age cut-off value`))[1]
store[loc,"Age cut-off value"] = i
# cut sample
Youngdata = HCP[which(HCP$Age <= i),]
# calculate correlations
## ICVerence
store[loc,"Cor"] =
with(Youngdata, cor.test(Age, ICV))$estimate
store[loc,"p"] =
with(Youngdata, cor.test(Age, ICV))$p.value
store[loc,"ci_l"] =
with(Youngdata, cor.test(Age, ICV))$conf.int[1]
store[loc,"ci_u"] =
with(Youngdata, cor.test(Age, ICV))$conf.int[2]
store[loc,"Measure"] = "ICV"
}
ggplot(data = store)+
geom_point(aes(x = Cor, y = `Age cut-off value`), alpha = 0.5)+
geom_errorbar(aes(y = `Age cut-off value`, xmin = ci_l, xmax = ci_u), alpha = 0.3)+
geom_vline(xintercept = -0.1, color = "grey")+
geom_hline(yintercept = 31, color = "grey")+
xlab("Correlation with age")+
ylab("Age cut-off used to subset\nsample into a younger subsample")+
scale_y_continuous(limits = c(21, 37.5), breaks = seq(from = 20, to = 36, by = 2))+
ggtitle("Age-ICV correlation in HCP")+
theme_bw()+
theme(panel.border = element_blank())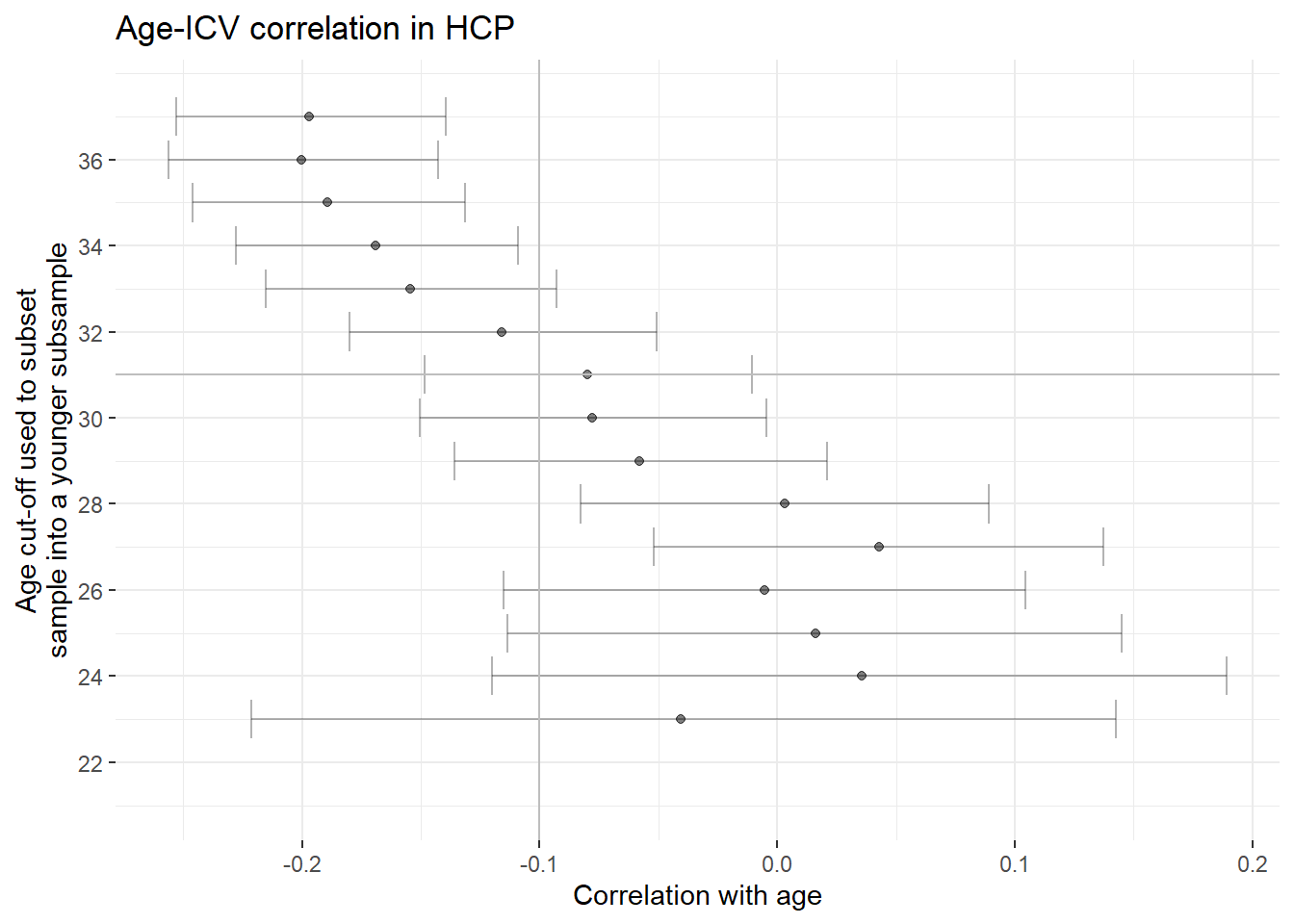
HCPyoung = HCP[which(HCP$Age <= 31),]
# calculate atrophy scores
HCPyoung$diff = HCPyoung$ICV - HCPyoung$TBV
HCPyoung$ratio = HCPyoung$TBV / HCPyoung$ICV
# delete those from data
#HCPyoung=HCPyoung[-which(HCPyoung$diff < 0),]
# estimate residual model
model <- lm(TBV ~ ICV, data = HCPyoung)
HCPyoung$resid = as.vector(resid(model, na.rm=T))
corICV = ggplot(data = HCPyoung, aes(x = ICV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
corTBV = ggplot(data = HCPyoung, aes(x = TBV, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
theme(panel.border = element_blank())
cordiff = ggplot(data = HCPyoung, aes(x = diff, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Difference score")+
theme(panel.border = element_blank())
corratio = ggplot(data = HCPyoung, aes(x = ratio, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Ratio score")+
theme(panel.border = element_blank())
corresid = ggplot(data = HCPyoung, aes(x = resid, y = Age))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
stat_cor(method = "pearson",color ="blue")+
ylab("Age in years")+
xlab("Residual score")+
theme(panel.border = element_blank())
ggarrange(corICV, corTBV, cordiff, corratio, corresid, common.legend=T, legend = "bottom")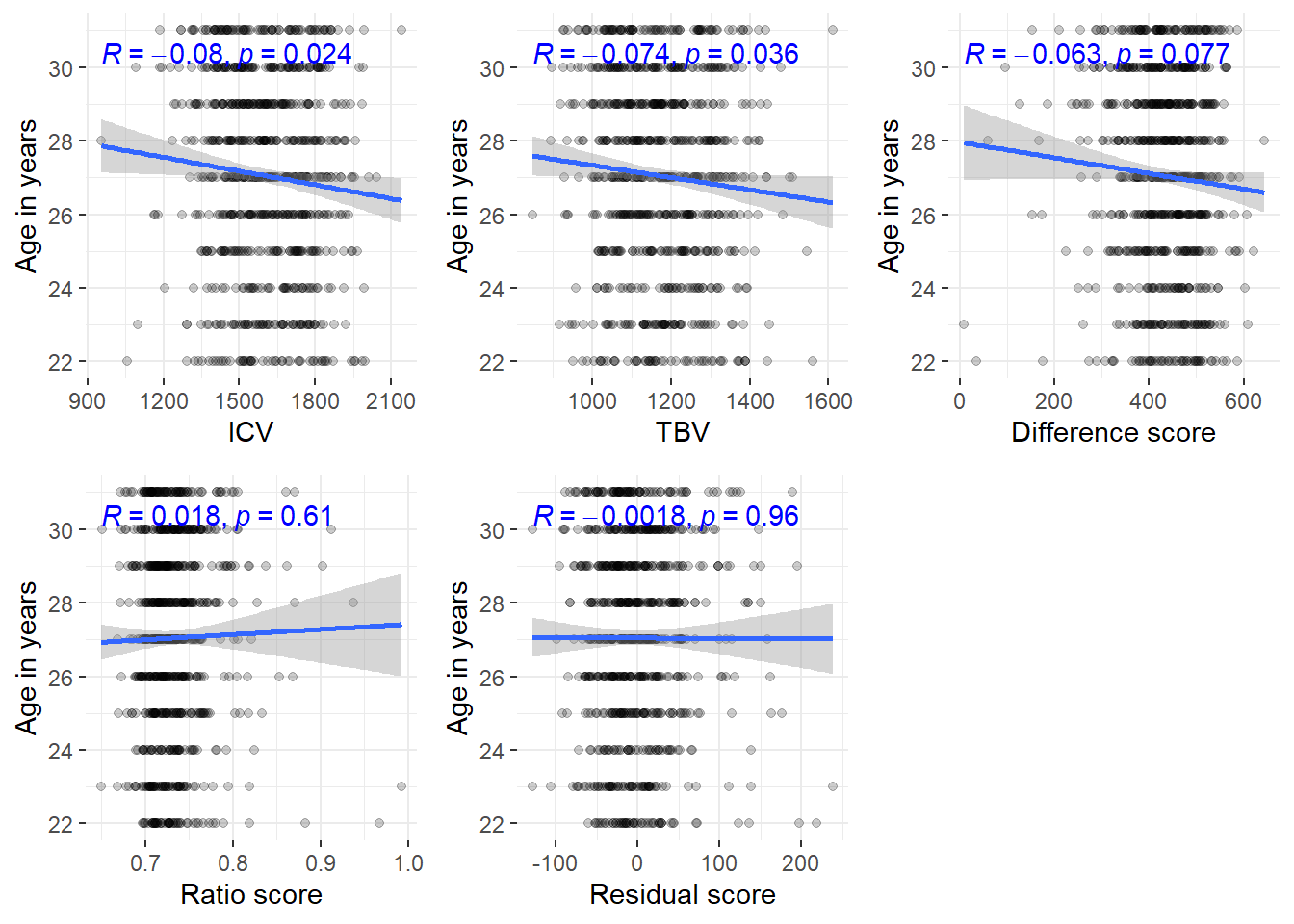
Shown in Supplementary Figure 3: Distributions of TBV, ICV, and lifetime brain atrophy estimated with the residual, ratio, and difference method. Histograms are coloured by age groups.
####################################################
# read in HCP data
HCP = fread(paste0(HCPdir,"/unrestricted_hcp_freesurfer.csv"))
HCP = HCP[,c("Subject", "Gender", "FS_InterCranial_Vol", "FS_BrainSeg_Vol_No_Vent")]
names(HCP) = c("ID", "Sex", "ICV", "TBV")
# add age information
HCPage = fread(paste0(HCPdir, "/RESTRICTED_annafurtjes_12_14_2023_4_18_2.csv"))
names(HCPage)[which(names(HCPage) == "Subject")] = "ID"
names(HCPage)[which(names(HCPage) == "Age_in_Yrs")] = "Age"
HCP = merge(HCP, HCPage[,c("ID","Age")], by = "ID")
# as outlined elsewhere, empirical investigations warrant to use an age cut-off of 31 years in this sample
HCP = HCP[which(HCP$Age <= 31),]
# convert mm3 estimates to more intuitive cm3 estimates
HCP$ICV = HCP$ICV/1000
HCP$TBV = HCP$TBV/1000
# estimate brain atrophy from single MRI scan
HCP$diff = HCP$ICV - HCP$TBV
HCP$ratio = HCP$TBV / HCP$ICV
# Quality control:
#print(paste0("Some participants have negative difference scores and ratio scores > 1, which means that their ICV estimate is smaller than their TBV estimate. This must be an error as the skull always surrounds the brain. Those ", sum((HCP$diff < 0))," HCP participants were excluded from the data set."))
deletedHCP = sum(HCP$diff < 0)
# delete those from data
if(sum(HCP$diff < 0) != 0){HCP=HCP[-which(HCP$diff < 0),]}
# estimate residual model
model <- lm(TBV ~ ICV, data = HCP)
HCP$resid = as.vector(resid(model, na.rm=T))
# standardise variables
HCP$diff_stand = as.vector(scale(HCP$diff))
HCP$ratio_stand = as.vector(scale(HCP$ratio))
HCP$resid_stand = as.vector(scale(HCP$resid))
####################################################
# make age groups
HCP$Age_group <- NA
HCP$Age_group[HCP$Age < 24] <- "23 years and under"
HCP$Age_group[HCP$Age >= 24 & HCP$Age < 25] <- "24"
HCP$Age_group[HCP$Age >= 25 & HCP$Age < 26] <- "25"
HCP$Age_group[HCP$Age >= 26 & HCP$Age < 27] <- "26"
HCP$Age_group[HCP$Age >= 27 & HCP$Age < 28] <- "27"
HCP$Age_group[HCP$Age >= 28 & HCP$Age < 29] <- "28"
HCP$Age_group[HCP$Age >= 29] <- "29 years and over"
p1=ggplot(HCP, aes(x=TBV, fill=Age_group)) +
geom_histogram()+
scale_fill_manual("Age groups", values = c("#292f56", "#1e4572", "#005c8b", "#008ba0", "#00bca1","#69e882", "#acfa70"))+
xlab("TBV")+
theme_bw()
p2=ggplot(HCP, aes(x=ICV, fill=Age_group)) +
geom_histogram()+
scale_fill_manual("Age groups", values = c("#292f56", "#1e4572", "#005c8b", "#008ba0", "#00bca1","#69e882", "#acfa70"))+
xlab("ICV")+
theme_bw()
p3=ggplot(HCP, aes(x=resid_stand, fill=Age_group)) +
geom_histogram()+
scale_fill_manual("Age groups", values = c("#292f56", "#1e4572", "#005c8b", "#008ba0", "#00bca1","#69e882", "#acfa70"))+
xlab("Residual score")+
theme_bw()
p4=ggplot(HCP, aes(x=ratio_stand, fill=Age_group)) +
geom_histogram()+
scale_fill_manual("Age groups", values = c("#292f56", "#1e4572", "#005c8b", "#008ba0", "#00bca1","#69e882", "#acfa70"))+
xlab("Ratio score")+
theme_bw()
p5=ggplot(HCP, aes(x=diff_stand, fill=Age_group)) +
geom_histogram()+
scale_fill_manual("Age groups", values = c("#292f56", "#1e4572", "#005c8b", "#008ba0", "#00bca1","#69e882", "#acfa70"))+
xlab("Difference score")+
theme_bw()
pHCP <- ggarrange(p1,p2,p3,p4,p5, nrow = 1, common.legend = T, legend = "bottom")
# add title
pHCP <- annotate_figure(pHCP, top = text_grob("HCP",face = "bold", size = 14))
#ggsave(paste0(out,"phenotypic/HCP_disttributions.jpg"), bg = "white",plot = pHCP, width = 30, height = 10, units = "cm", dpi = 300)
pHCP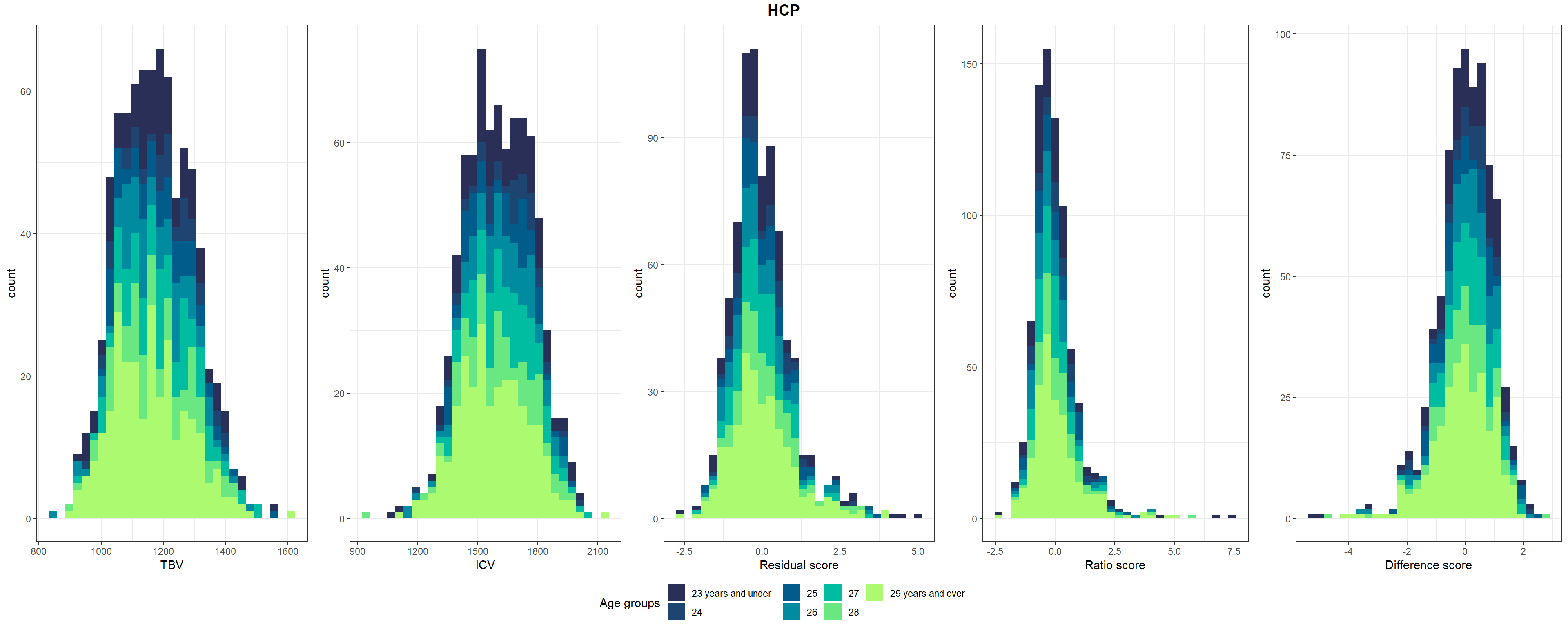
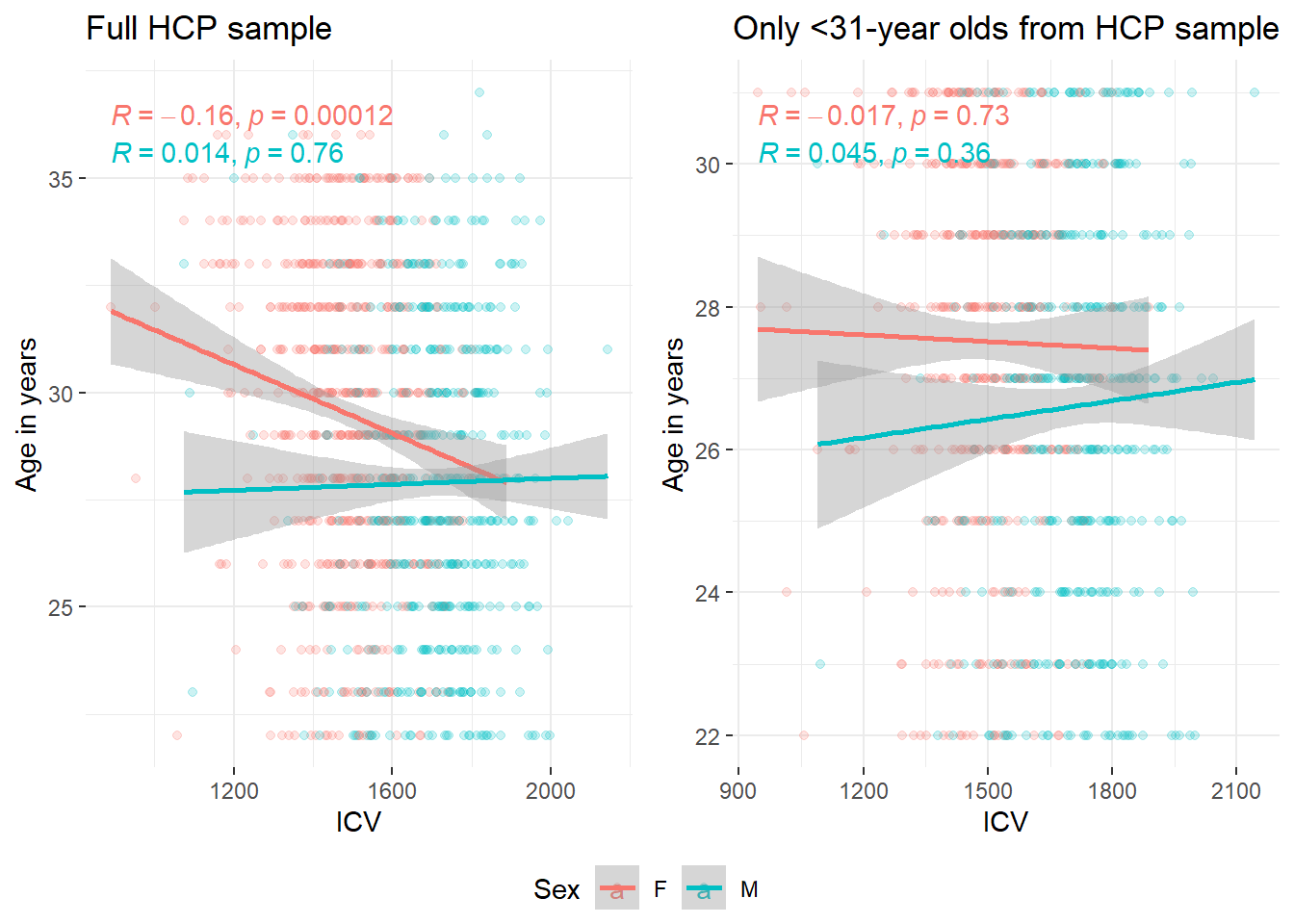
It was brought to our attention through the revision process of this project that the HCP has been shown to over-represent males at younger ages and that there are more females at older ages, which could also explain this effect here.
The plots below show a clear sex difference in ICV and TBV, as expected (i.e., larger ICV in males). It also shows that the unusual direction of effects (smaller heads at older ages) is driven by females.
HCPcov = fread(paste0(HCPdir,list.files(path = HCPdir, pattern = "unrestricted_annafurtjes")))
HCPcov = HCPcov[,c("Subject", "Release", "Acquisition","fMRI_3T_ReconVrs")]
HCP = fread(paste0(HCPdir,"unrestricted_hcp_freesurfer.csv"))
HCP = HCP[,c("Subject", "Gender", "FS_InterCranial_Vol", "FS_BrainSeg_Vol_No_Vent")]
HCP = merge(HCP, HCPcov, by = "Subject")
HCPage = fread(paste0(HCPdir, "RESTRICTED_annafurtjes_12_14_2023_4_18_2.csv"))
HCPage = HCPage[,c("Subject","Age_in_Yrs", "Family_ID")]
HCP = merge(HCP, HCPage, by ="Subject")
names(HCP) = c("ID", "Sex", "ICV", "TBV", "Release", "Acquisition","fMRI_3T_ReconVrs", "Age", "Family_ID")
# convert mm3 estimates to more intuitive cm3 estimates
HCP$ICV = HCP$ICV/1000
HCP$TBV = HCP$TBV/1000
# calculate atrophy scores
HCP$diff = HCP$ICV - HCP$TBV
HCP$ratio = HCP$TBV / HCP$ICV
# delete those from data
HCP=HCP[-which(HCP$diff < 0),]
# estimate residual model
model <- lm(TBV ~ ICV, data = HCP)
HCP$resid = as.vector(resid(model, na.rm=T))
# standardise variables
HCP$diff_stand = as.vector(scale(HCP$diff))
HCP$ratio_stand = as.vector(scale(HCP$ratio))
HCP$resid_stand = as.vector(scale(HCP$resid))
corICV = ggplot(data = HCP, aes(x = ICV, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
stat_cor(aes(color = Sex), method = "pearson")+
theme_bw()+
geom_smooth(method='lm')+
ylab("Age in years")+
theme(panel.border = element_blank())+
ggtitle("Full HCP sample")
corTBV = ggplot(data = HCP, aes(x = TBV, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
stat_cor(aes(color = Sex), method = "pearson")+
theme_bw()+
geom_smooth(method='lm')+
ylab("Age in years")+
theme(panel.border = element_blank())
cordiff = ggplot(data = HCP, aes(x = diff, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
theme_bw()+
stat_cor(aes(color = Sex), method = "pearson")+
geom_smooth(method='lm')+
ylab("Age in years")+
xlab("Difference score")+
theme(panel.border = element_blank())
corratio = ggplot(data = HCP, aes(x = ratio, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
theme_bw()+
geom_smooth(method='lm')+
ylab("Age in years")+
xlab("Ratio score")+
theme(panel.border = element_blank())
corresid = ggplot(data = HCP, aes(x = resid, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
stat_cor(aes(color = Sex), method = "pearson")+
theme_bw()+
geom_smooth(method='lm')+
ylab("Age in years")+
xlab("Residual score")+
theme(panel.border = element_blank())
ggarrange(corICV, corTBV, cordiff, corratio, corresid, common.legend=T, legend = "bottom")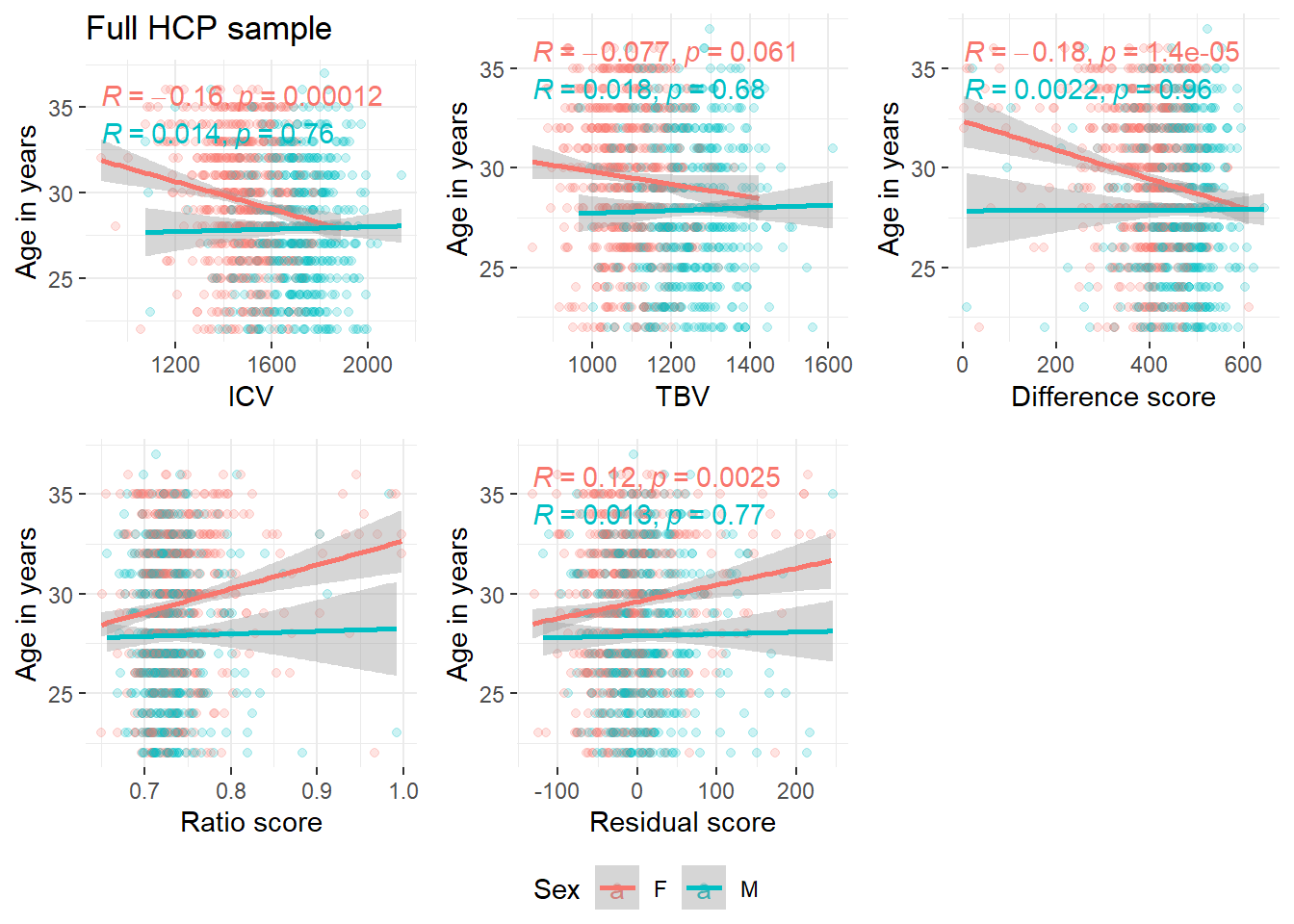
####################################################
# read in HCP data
HCP = fread(paste0(HCPdir,"/unrestricted_hcp_freesurfer.csv"))
HCP = HCP[,c("Subject", "Gender", "FS_InterCranial_Vol", "FS_BrainSeg_Vol_No_Vent")]
names(HCP) = c("ID", "Sex", "ICV", "TBV")
# add age information
HCPage = fread(paste0(HCPdir, "/RESTRICTED_annafurtjes_12_14_2023_4_18_2.csv"))
names(HCPage)[which(names(HCPage) == "Subject")] = "ID"
names(HCPage)[which(names(HCPage) == "Age_in_Yrs")] = "Age"
HCP = merge(HCP, HCPage[,c("ID","Age")], by = "ID")
# convert mm3 estimates to more intuitive cm3 estimates
HCP$ICV = HCP$ICV/1000
HCP$TBV = HCP$TBV/1000
# estimate brain atrophy from single MRI scan
HCP$diff = HCP$ICV - HCP$TBV
HCP$ratio = HCP$TBV / HCP$ICV
# Quality control:
#print(paste0("Some participants have negative difference scores and ratio scores > 1, which means that their ICV estimate is smaller than their TBV estimate. This must be an error as the skull always surrounds the brain. Those ", sum((HCP$diff < 0))," HCP participants were excluded from the data set."))
deletedHCP = sum(HCP$diff < 0)
# delete those from data
if(sum(HCP$diff < 0) != 0){HCP=HCP[-which(HCP$diff < 0),]}
# estimate residual model for males and females separately
model <- lm(TBV ~ ICV, data = HCP)
HCP$resid = as.vector(resid(model, na.rm=T))
# estimate residual model for males and females separately
# males
#males <- HCP[HCP$Sex == "M",]
#model <- lm(TBV ~ ICV, data = males)
#males$resid = as.vector(resid(model, na.rm=T))
#females
#females <- HCP[HCP$Sex == "F",]
#model <- lm(TBV ~ ICV, data = females)
#females$resid = as.vector(resid(model, na.rm=T))
# merge males and females back together
#both = rbind(males,females)
# ICV
icv = ggplot(HCP, aes(y = ICV, x = Sex, fill = Sex))+
geom_boxplot(alpha = 0.6, color = "black")+
geom_point(aes(fill = factor(Sex)),
color = "grey45",
size = 2,
alpha = 0.5,
position = position_jitter(seed = 1, width = 0.2))+
xlab("Sex")+
ylab("ICV")+
theme_bw()+
theme(legend.position = "none")+
theme(text = element_text(size=20),
axis.text.x = element_text(size=20),#angle=45
axis.text.y = element_text(size=20),
axis.title.y = element_text(colour='black', size=20),
axis.title.x = element_text(colour='black', size=20),
plot.title = element_text(face = "bold", colour='black', size=20))
tbv = ggplot(HCP, aes(y = TBV, x = Sex, fill = Sex))+
geom_boxplot(alpha = 0.6, color = "black")+
geom_point(aes(fill = factor(Sex)),
color = "grey45",
size = 2,
alpha = 0.5,
position = position_jitter(seed = 1, width = 0.2))+
xlab("Sex")+
ylab("TBV")+
theme_bw()+
theme(legend.position = "none")+
theme(text = element_text(size=20),
axis.text.x = element_text(size=20),#angle=45
axis.text.y = element_text(size=20),
axis.title.y = element_text(colour='black', size=20),
axis.title.x = element_text(colour='black', size=20),
plot.title = element_text(face = "bold", colour='black', size=20))
# residual score
res = ggplot(HCP, aes(y = resid, x = Sex, fill = Sex))+
geom_boxplot(alpha = 0.6, color = "black")+
geom_point(aes(fill = factor(Sex)),
color = "grey45",
size = 2,
alpha = 0.5,
position = position_jitter(seed = 1, width = 0.2))+
xlab("Sex")+
ylab("Residual score (standardised)\n<- More brain atrophy Less brain atrophy ->")+
scale_fill_manual(values = c("#004225", "#90ee90"))+
theme_bw()+
theme(legend.position = "none")+
theme(text = element_text(size=20),
axis.text.x = element_text(size=20),#angle=45
axis.text.y = element_text(size=20),
axis.title.y = element_text(colour='black', size=20),
axis.title.x = element_text(colour='black', size=20),
plot.title = element_text(face = "bold", colour='black', size=20))
# ratio score
rat = ggplot(HCP, aes(y = ratio, x = Sex, fill = Sex))+
geom_boxplot(alpha = 0.6, color = "black")+
geom_point(aes(fill = factor(Sex)),
color = "grey45",
size = 2,
alpha = 0.5,
position = position_jitter(seed = 1, width = 0.2))+
xlab("Sex")+
ylab("Ratio score (standardised)\n<- More brain atrophy Less brain atrophy ->")+
scale_fill_manual(values = c("#ffc40c", "#fcffa4"))+
theme_bw()+
theme(legend.position = "none")+
theme(text = element_text(size=20),
axis.text.x = element_text(size=20),#angle=45
axis.text.y = element_text(size=20),
axis.title.y = element_text(colour='black', size=20),
axis.title.x = element_text(colour='black', size=20),
plot.title = element_text(face = "bold", colour='black', size=20))
# difference score
dif = ggplot(HCP, aes(y = diff, x = Sex, fill = Sex))+
geom_boxplot(alpha = 0.6, color = "black")+
geom_point(aes(fill = factor(Sex)),
color = "grey45",
size = 2,
alpha = 0.5,
position = position_jitter(seed = 1, width = 0.2))+
xlab("Sex")+
ylab("Ratio score (standardised)\n<- More brain atrophy Less brain atrophy ->")+
scale_fill_manual(values = c("#ff1493", "#ffb6c1"))+
theme_bw()+
theme(legend.position = "none")+
theme(text = element_text(size=20),
axis.text.x = element_text(size=20),#angle=45
axis.text.y = element_text(size=20),
axis.title.y = element_text(colour='black', size=20),
axis.title.x = element_text(colour='black', size=20),
plot.title = element_text(face = "bold", colour='black', size=20))
plot_grid(icv, tbv, nrow = 1)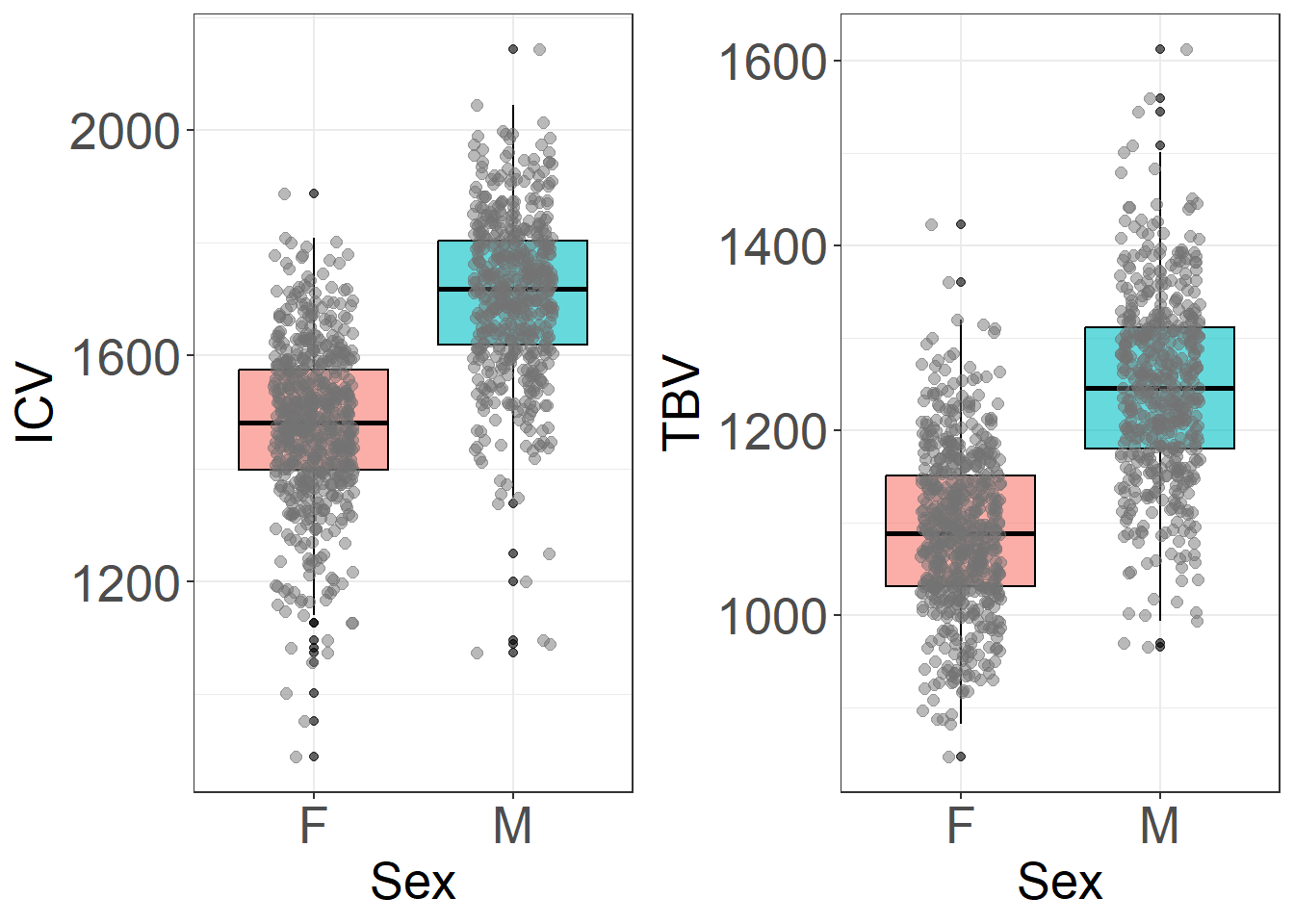
Test if sex effect is still there when restricting participants to <31 year-olds.
# read in HCP data
HCP = fread(paste0(HCPdir,"/unrestricted_hcp_freesurfer.csv"))
HCP = HCP[,c("Subject", "Gender", "FS_InterCranial_Vol", "FS_BrainSeg_Vol_No_Vent")]
names(HCP) = c("ID", "Sex", "ICV", "TBV")
# add age information
HCPage = fread(paste0(HCPdir, "/RESTRICTED_annafurtjes_12_14_2023_4_18_2.csv"))
names(HCPage)[which(names(HCPage) == "Subject")] = "ID"
names(HCPage)[which(names(HCPage) == "Age_in_Yrs")] = "Age"
HCP = merge(HCP, HCPage[,c("ID","Age")], by = "ID")
# convert mm3 estimates to more intuitive cm3 estimates
HCP$ICV = HCP$ICV/1000
# as outlined elsewhere, empirical investigations warrant to use an age cut-off of 31 years in this sample
HCP = HCP[which(HCP$Age <= 31),]
#corICV
corICV_below31= ggplot(data = HCP, aes(x = ICV, y = Age, color = Sex))+
geom_point(alpha = 0.2)+
stat_cor(aes(color = Sex), method = "pearson")+
theme_bw()+
geom_smooth(method='lm')+
ylab("Age in years")+
theme(panel.border = element_blank())+
ggtitle("Only <31-year olds from HCP sample")
ggarrange(corICV, corICV_below31, common.legend = T, legend = "bottom")`geom_smooth()` using formula = 'y ~ x'
`geom_smooth()` using formula = 'y ~ x'
`geom_smooth()` using formula = 'y ~ x'